windowdiversity¶
Result = windowdiversity(expression, windowlength)
- expression
spatial boolean, nominal, ordinal
- windowlength
spatial, non spatial scalar
- Result
spatial scalar
Options¶
--unittrue or --unitcell
--unittruewindowlength is measured in true length (default)
--unitcellwindowlength is measured in number of cell lengths
Operation¶
For each cell a square window with the cell in its centre is defined by windowlength. The windowlength is the length of the window in horizontal and vertical directions. For each cell on expression the number of unique values within its window is counted. This number is assigned to the corresponding cell on Result. Both cells on expression which are entirely in the window and cells which are partly in the window are considered.
Notes¶
The cell value on windowlength should be greater than 0, else a missing value is assigned to the corresponding cell on Result.
A cell on windowlength with a missing value results in a missing value on Result at the corresponding cell. However, if a missing value on windowlength occurs in the window in a cell which is not the centre cell of the window the expression value in that cell is considered for determination of the number of unique values in the window.
Group¶
This operation belongs to the group of Neigbourhood operators; window operators
Examples¶
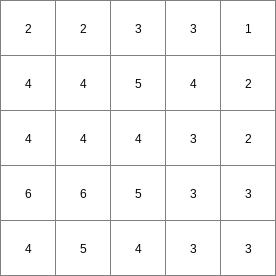
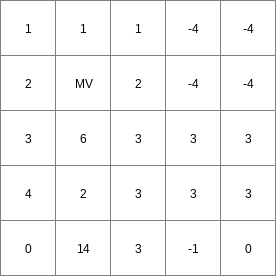
- • pcrcalcbindingResult2 = Result2.map;Expr = Expr.map;WinLen2 = WinLen2.map;initialreport Result2 = windowdiversity( Expr, WinLen2);• pythonExpr = readmap(“Expr.map”)WinLen2 = readmap(“WinLen2.map”)Result2 = windowdiversity( Expr, WinLen2)
Result2.map
Expr.map
WinLen2.map

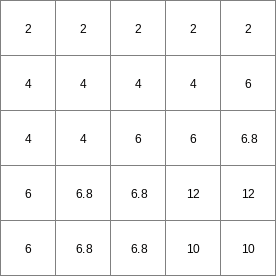
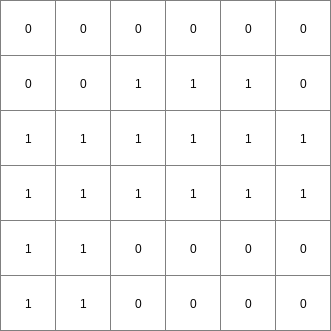
- • pcrcalcbindingResult3 = Result3.map;Expr2 = Expr2.map;initialreport Result3 = (windowdiversity(Expr2,celllength() * 1.1)) > 1;• pythonExpr2 = readmap(“Expr2.map”)Result3 = (windowdiversity(Expr2,celllength() * 1.1)) > 1
Result3.map
Expr2.map
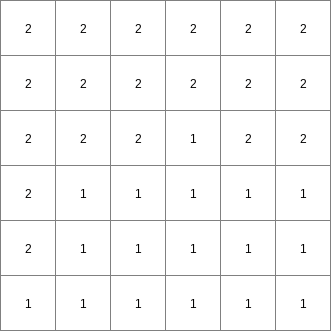
- • pcrcalcbindingResult1 = Result1.map;Expr = Expr.map;initialreport Result1 = windowdiversity( Expr, 6);• pythonExpr = readmap(“Expr.map”)Result1 = windowdiversity( Expr, 6)
Result1.map
Expr.map