path¶
Result = path(ldd, points)
- ldd
spatial ldd
- points
spatial boolean
- Result
spatial boolean
Operation¶
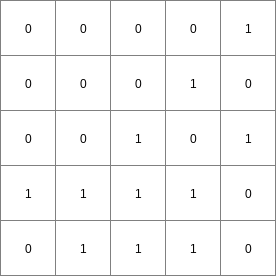
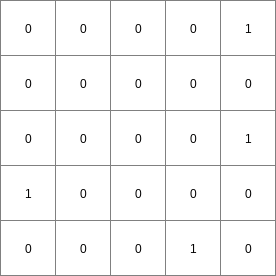
The cell values on points are interpreted as Boolean values; where 1 is TRUE and 0 is FALSE. The operation determines for each TRUE cell on points the cells which are on the path downstream to its pit. Each path is generated by starting at the TRUE cell on points and moving through the consecutively neighbouring downstream cells, following the local drain directions on ldd. On Result, all cells which are on the path of one or more TRUE cells on points are assigned a 1 (TRUE), the cells which are not on a path are assigned a 0 (FALSE).
Notes¶
A missing value on points and/or ldd results in a missing value at the corresponding cell on Result. A path stops at a missing value cell on points. Cells on the downstream path of a missing value on points are assigned a 0, unless they are on another path from a TRUE cell on points.
Group¶
This operation belongs to the group of Neighbourhood operators; local drain directions
See Also¶
Examples¶
- • pcrcalcbindingResult = Result.map;Ldd = Ldd.map;Points = Points.map;initialreport Result = path(Ldd,Points);• pythonLdd = readmap(“Ldd.map”)Points = readmap(“Points.map”)Result = path(Ldd,Points)
Result.map
Ldd.map
Points.map