accuthresholdflux, accuthresholdstate¶
Resultflux = accuthresholdflux(ldd, material, transportthreshold)
Resultstate = accuthresholdstate(ldd, material, transportthreshold)
Resultflux Resultstate = accuthresholdflux, accuthresholdstate(ldd, material, transportthreshold)
- ldd
spatial ldd
- transportthreshold
spatial, non spatial scalar
- material
spatial, non spatial scalar
- Resultstate
spatial scalar
- Resultflux
spatial scalar
Operation¶
These operations describe accumulation of material in a drainage network with transport limited by a threshold: transport will only occur if a certain threshold of losses has been reached. Material less than the threshold is stored. This is the case for overland flow which will only develop once a certain loss has occurred, saturating the soil. The mechanism can also be used to describe phenomena such as losses from the streamflow due to infiltration of river water through the riverbed.
For each cell, accuthresholdflux assigns the amount of material which is transported out of the cell, accuthresholdstate assigns the amount which is stored in the cell. Both operators perform the same function of accumulation of material with a transport threshold, the only difference between the operators is the sort of result that is saved: accuthresholdstate yields storages of material in cells, accuthresholdflux yields fluxes of material out of cells.
For each cell, the amount of material input, for instance the amount of rain, is given by material. This is transported in downstream direction through the consecutively neighbouring downstream cells, following the local drain directions on ldd. Each time material moves through a cell an certain amount is stored in the cell. These storages are saved as Resultstate, if the accuthresholdstate operator is used. The remaining material is transported out of the cell, these amounts of outflow from each cell into its neighbouring downstream cell are the result of the accuthresholdflux operator, they are saved as Resultflux.
The function can be described by flow of material through a set of linked systems, where a cell represents a system. The flow starts at the cells/systems at the watershed boundaries (defined by ldd) and ends at a pit cell. The systems are linked by the local drain directions on ldd, these define the path of flow through the set of cells/systems. Each time a system is passed, the amount of flow changes.
For a cell/system somewhere on the map, the flow of material is described by a system. The inflow of the cell is the sum of the outflow amounts of its upstream neighbours. This inflow amount is added to the material value in the cell itself. This amount of material is potentially available for transport out of the cell. If it is less than or equal to the transportthreshold value of the cell all material is stored. If it is more than the transportthreshold the amount transported is the amount potentially available for transport minus the transportthreshold value. The remaining material is stored.
For each cell, the amount of material which is transported to its downstream neighbour (or out of the map if the cell is a pit cell) is saved as Resultflux (use the operator accuthresholdflux); the amount of material which is stored to the cell is saved as Resultstate (use accuthresholdstate)
Notes¶
The values on material and transportthreshold must be equal to or larger than zero.
A cell with missing value on material and/or transportthreshold is assigned a missing value on Resultflux or Resultstate. Additionally, all its downstream cells are assigned a missing value.
The local drain direction network on ldd must be sound.
Group¶
This operation belongs to the group of Neighbourhood operators; local drain directions
See Also¶
Examples¶
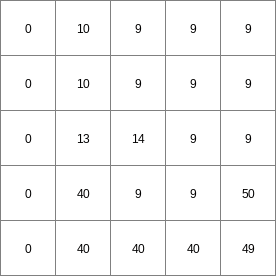
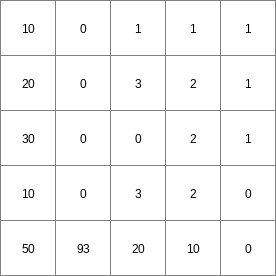
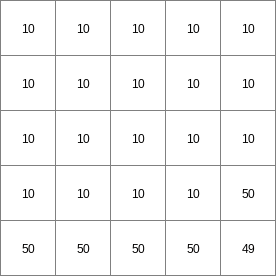
- • pcrcalcbindingState1 = State1.map;Flux1 = Flux1.map;Ldd = Ldd.map;Material = Material.map;initialreport State1, Flux1 = accuthresholdstate,accuthresholdflux(Ldd,Material,1.5);• pythonLdd = readmap(“Ldd.map”)Material = readmap(“Material.map”)State1 = accuthresholdstate(Ldd,Material,1.5)Flux1 = accuthresholdflux(Ldd,Material,1.5)
State1.map
Flux1.map
Ldd.map
Material.map
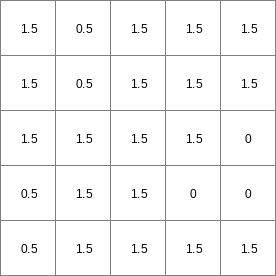
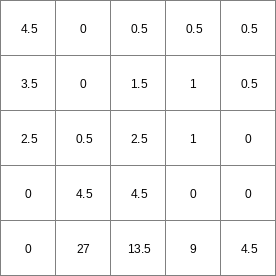

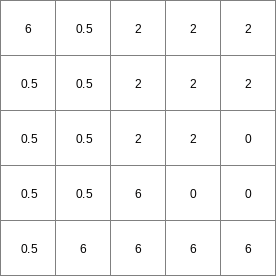
- • pcrcalcbindingState2 = State2.map;Flux2 = Flux2.map;Ldd = Ldd.map;Material = Material.map;TransTH = TransTH.map;initialreport State2, Flux2 = accuthresholdstate,accuthresholdflux(Ldd,Material,TransTH);• pythonLdd = readmap(“Ldd.map”)Material = readmap(“Material.map”)TransTH = readmap(“TransTH.map”)State2 = accuthresholdstate(Ldd,Material,TransTH)Flux2 = accuthresholdflux(Ldd,Material,TransTH)
State2.map
Flux2.map
Ldd.map
Material.map
TransTH.map